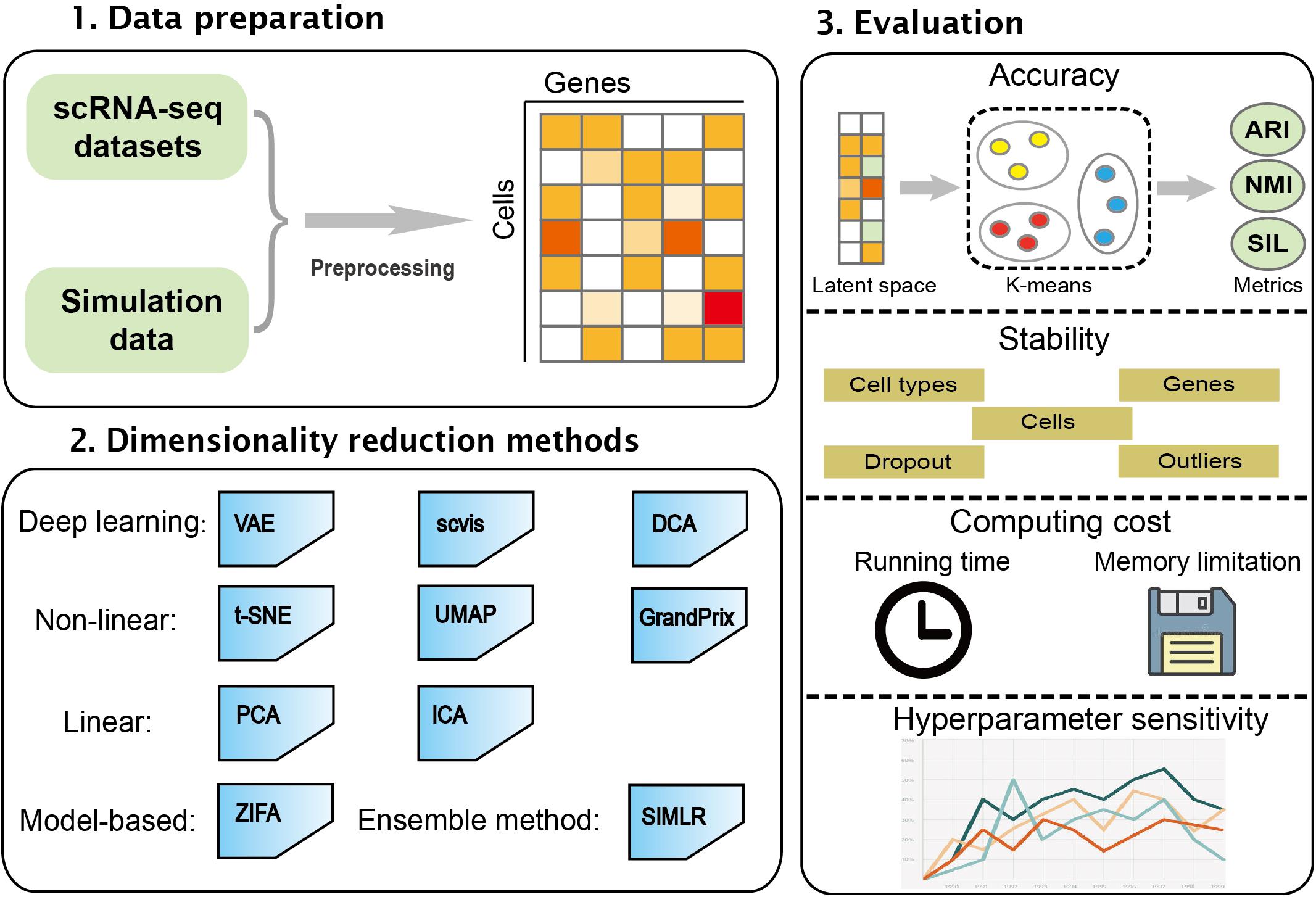
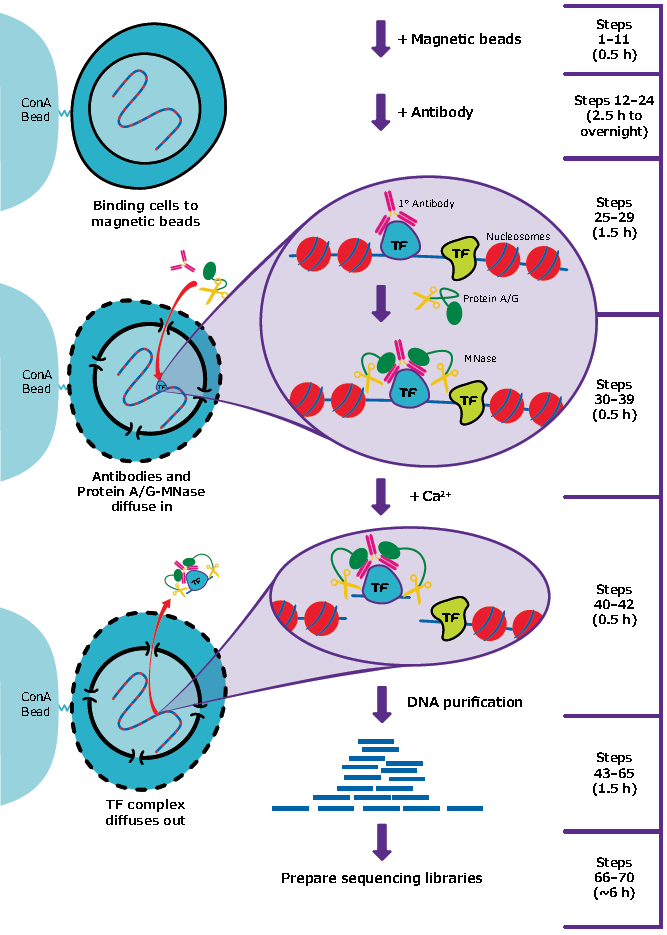
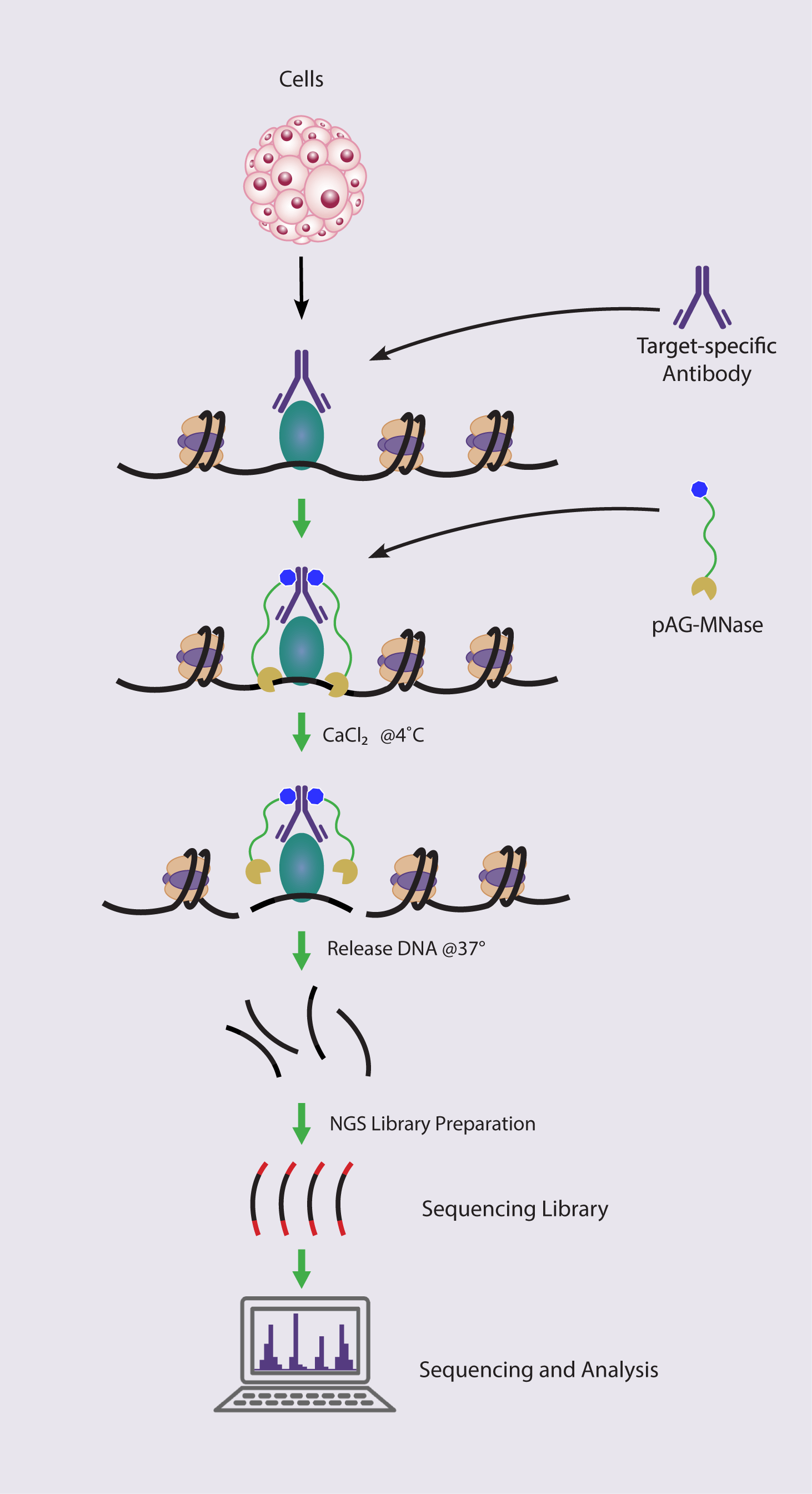
New Chromatin Run-On Reaction Enables Global Mapping of Active RNA Polymerase Locations in an Enrichment-free Manner | ACS Chemical Biology

Protocol variations in run-on transcription dataset preparation produce detectable signatures in sequencing libraries | BMC Genomics | Full Text

Genomics for antimicrobial resistance surveillance to support infection prevention and control in health-care facilities - The Lancet Microbe

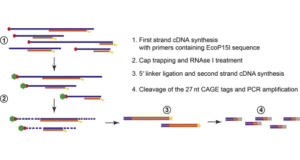
Principles and innovative technologies for decrypting noncoding RNAs: from discovery and functional prediction to clinical application | Journal of Hematology & Oncology | Full Text

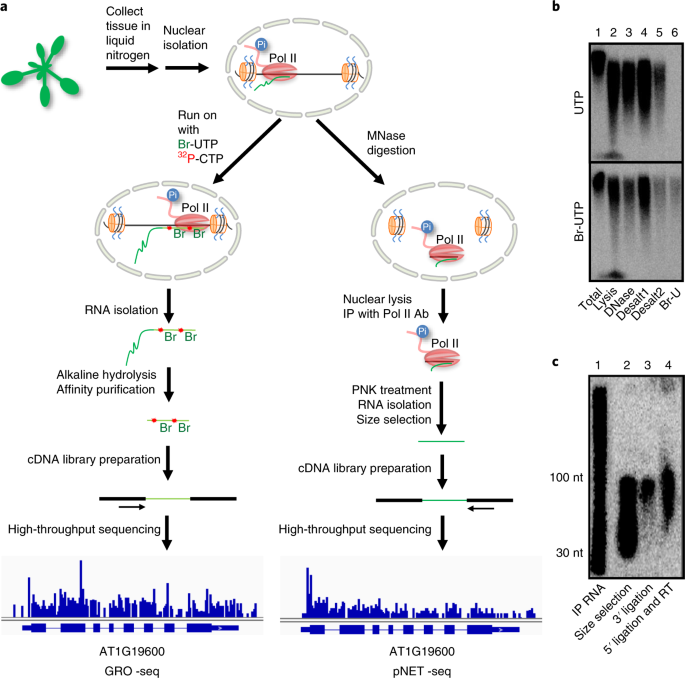
Global nuclear run-on deep sequencing (GRO-seq) indicates that Pol II... | Download Scientific Diagram
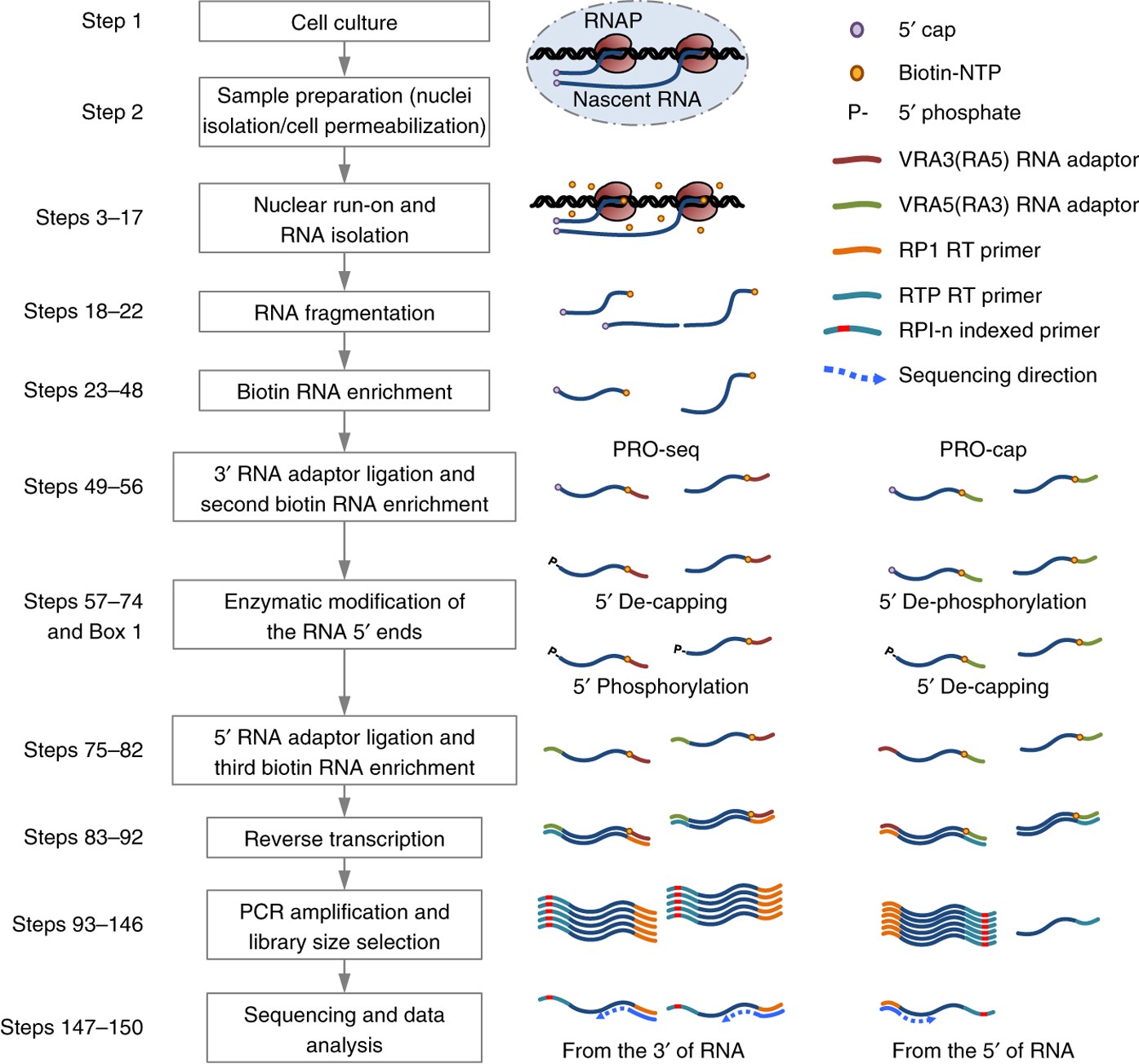
Base-pair-resolution genome-wide mapping of active RNA polymerases using precision nuclear run-on (PRO-seq) | Nature Protocols

Full article: A comparison of existing global DNA methylation assays to low-coverage whole-genome bisulfite sequencing for epidemiological studies

New Chromatin Run-On Reaction Enables Global Mapping of Active RNA Polymerase Locations in an Enrichment-free Manner | ACS Chemical Biology
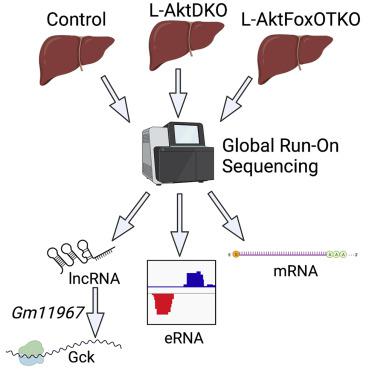
Global-run on sequencing identifies Gm11967 as an Akt-dependent long noncoding RNA involved in insulin sensitivity,iScience - X-MOL

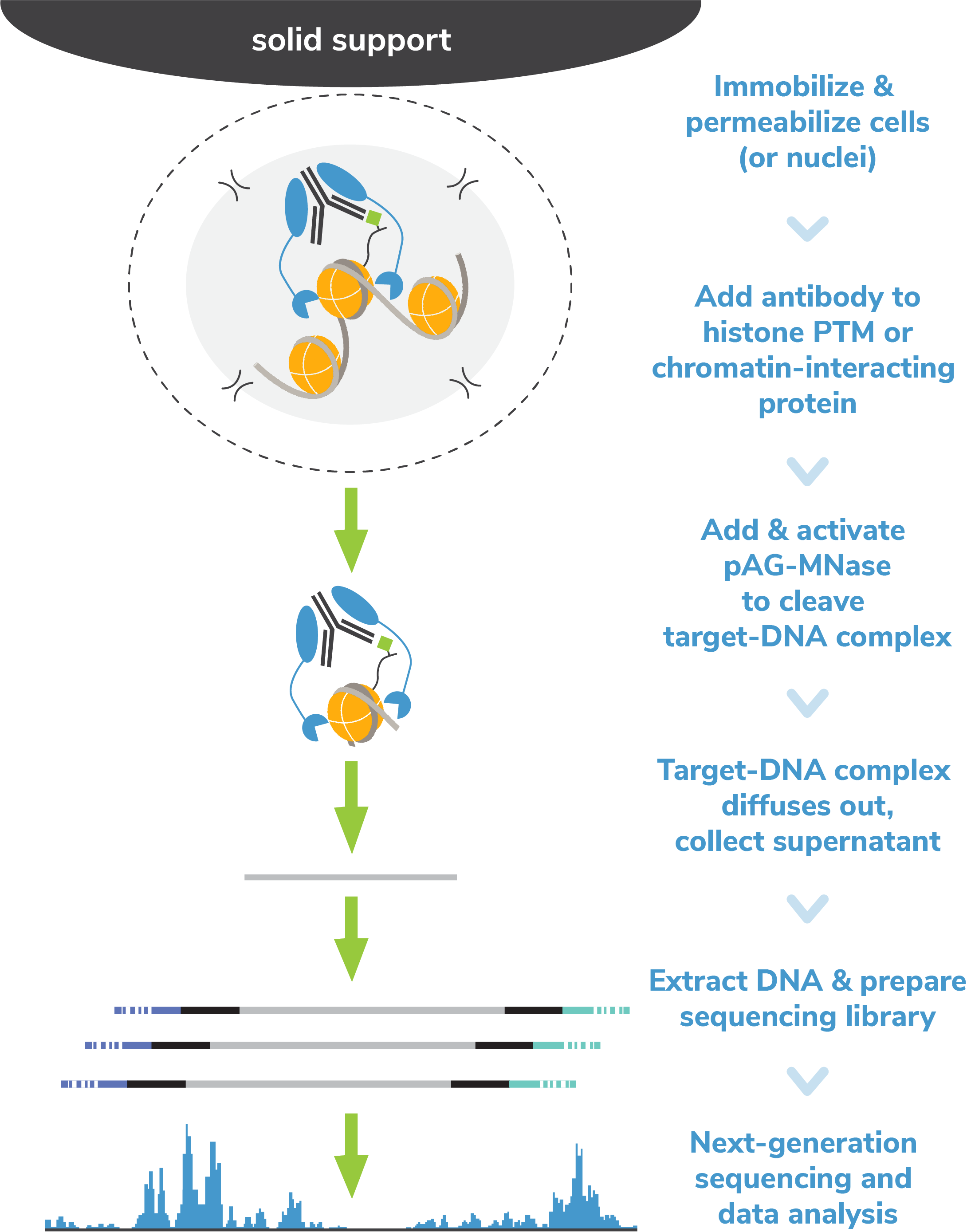
Native Elongating Transcript Sequencing Reveals Human Transcriptional Activity at Nucleotide Resolution - ScienceDirect

Fast Read Stitcher – an annotation agnostic algorithm for detecting nascent RNA transcripts in global nuclear run-on sequencing (GRO-seq) | RNA-Seq Blog

Global patterns of antigen receptor repertoire disruption across adaptive immune compartments in COVID-19 | PNAS